|
|
Welcome to the Mutwil Lab!
The Mutwil lab is located within the School of Biological Sciences at Nanyang Technological University, Singapore.
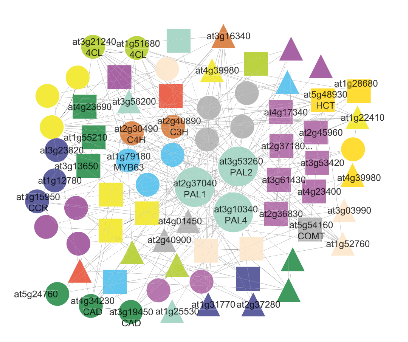
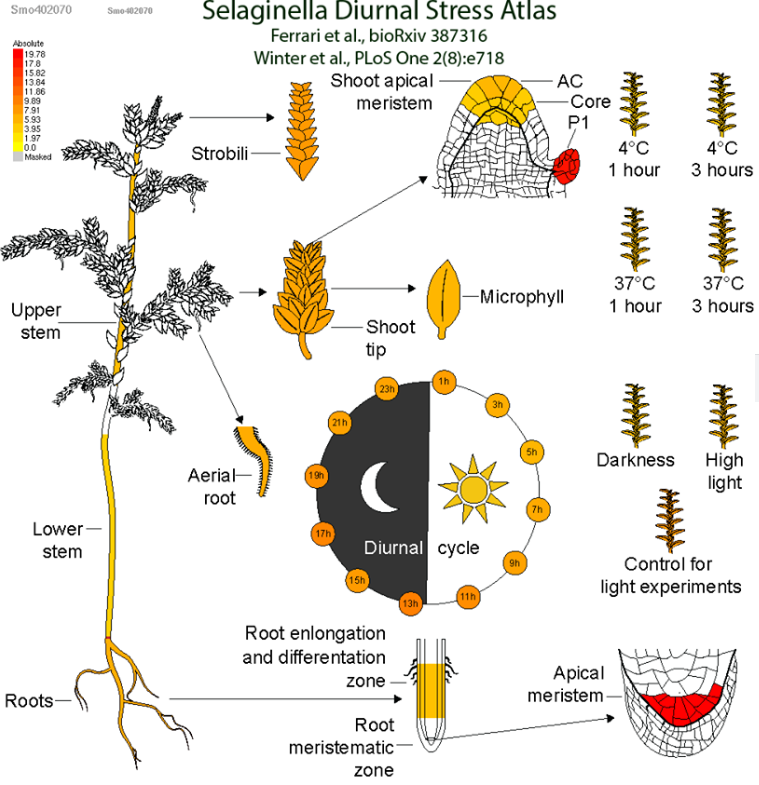
Our group aims to uncover the function and evolution of genes, stress resilience mechanisms, and specialized metabolic biosynthetic pathways in algae and land plants using large-scale, comparative experimental, and computational approaches. Our work focuses on four interconnected research pillars: Evolution, Stress Resilience, Specialized Metabolism, and Tools & Databases. By analyzing genomic, expression, and protein-protein interaction data for hundreds of plant species, the research will expand knowledge of gene function and the evolution of the plant kingdom, with applications in engineering stress-resilient crops with higher nutritional values. The team's findings will be made accessible through online databases, which will offer invaluable resources to researchers studying gene function, evolution, and stress resilience in plants.















 RSS Feed
RSS Feed